Researchers at the Department of Ophthalmology, at The Hebrew University of Jerusalem, have reported a new comprehensive analysis on Best disease patients, aimed to document mutation analyses, genotype–phenotype correlations, and an estimate of disease prevalence in the Israeli population. Best vitelliform macular dystrophy, known as Best disease, is a progressive macular degeneration, first described in 1905, and is caused by heterozygous mutations in the gene “BEST1” (VMD2, vitelliform macular dystrophy-2). The researchers commented that, based on these findings, “we were able to calculate the maximum and the minimum prevalence of Best disease in the Israeli population and different subpopulations that reside in Israel. In addition, after reviewing previously published papers, it appears that mutations in BEST1 can cause a broad clinical spectrum of macular dystrophies, which suggests a multifunctional role of the protein in the retina.”
The researchers collected medical records of patients diagnosed with Best disease from nine Israeli medical centres, including clinical data from ocular findings, electrophysiology results, and retina imaging. Of the analysis, the researchers reported a total of 134 patients with Best disease and related conditions, estimating a prevalence to be 1 in 127,000, with higher rates among Arab Muslims (1 in 76,000) than Jews (1 in 145,000). Critical conserved domains were identified consisting of a high percentage of dominant missense mutations, primarily in transmembrane domains and the intracellular region (Ca2+ binding domain) of the BEST1 protein.
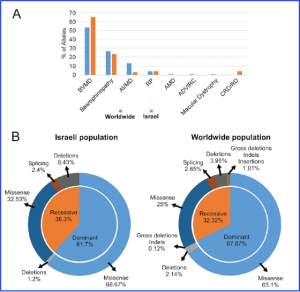
Figure 1 (A) The distribution of BEST1 mutated alleles in the studied set of Israeli patients compared to the alleles reported previously. The data were calculated and are presented in percent (%). Each bar represents the percentage of alleles associated with a certain clinical diagnosis among the Israeli cohort of BEST1 (orange) and worldwide (blue). (B) The distribution of mutated BEST1 alleles identified in patients from the Israeli cohort (left pie chart) and alleles reported worldwide (right pie chart) based on recessive/dominant alleles (inner circle) and the type of the mutation (outer circle). [Copyright from IOVS, under a Creative Commons license – Beryozkin et al., Investigative Ophthalmology & Visual Science February 2024, Vol.65, 39].
According to their publication, the researchers commented that, “to our knowledge, this is the largest cohort of patients with Best disease reported to date in the Israeli population and worldwide”. The information of these results may be extremely valuable which might benefit from the development of gene augmentation therapy or RNA editing therapy in due course. Their research team commented that “the findings from this research will enable us and other researchers to better understand the range of mutations that cause the disease, identify crucial functional domains, and explore possible correlations between genotypes and phenotypes. Ultimately, this knowledge will contribute to unravelling the molecular mechanisms underlying BVMD and guide the development of novel treatments”.