Researchers based in Moorfields Eye Hospital, NHS Foundation Trust, London, have presented the first genome-wide association study (GWAS) results involving reticular pseudodrusen (RPD). The study’s analysis provided a clear association between the ARMS2-HTRA1 locus and higher RPD load, including three novel associations unique to RPD. Given that the genetic basis of RPD is not as well characterised in the literature as preferred, the UK research team aimed to hypothesize “that RPD and drusen arise from both shared and distinct biological pathways.” The team used a genome-wide association study approach using retinal imaging data from the UK Biobank (UKBB) resource, leveraging a validated AI algorithm to identify and quantify drusen and RPD.
While drusen is normal with advancing age, and can be seen by small yellow or white accumulations of extracellular material that build up between Bruch’s membrane and the retinal pigment epithelium (RPE), reticular pseudodrusen appears more prominent under blue light. In contrary to drusen which locates below the RPE, reticular pseudodrusen is located superficial to the RPE, more commonly found at the superotemporal quadrant of the macula. According to the current authors’ GWAS study, “several lines of evidence highlight the distinctions between drusen and RPD, including their anatomic location (below and above the RPE, respectively); differences in biochemical composition; a significantly increased risk of progression in AMD patients with RPD compared to without; and appearance of RPD in conditions where drusen are not part of the main phenotype.”
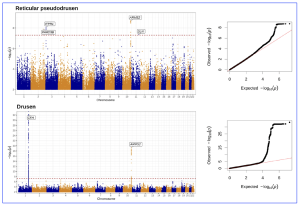
GWAS have been conducted since 2002 and there have been >4,000 studies conducted worldwide, reporting ~55,000 unique genetic associations across nearly ~5,000 traits and diseases. GWAS aim to identify single-nucleotide polymorphisms (SNPs) that occur more frequently within the genomes of a given disease population than in a control population and the SNPs can infer genomic regions for disease association. In the current study from Moorefield hospital, their results showed that the primary GWAS analyses on their AMD cohort yielded 4 loci reaching genome-wide significance for pure RPD (ARMS2-HTRA1, PARD3B, ITPR1 and SLN) and 2 loci for pure drusen (CFH and ARMS2-HTRA1), summarized in the figure below:
Figure 1. Manhattan and quantile-quantile plots of GWAS results for number of pure RPD (in the absence of drusen) and number of pure drusen (in the absence of RPD). [The research work is licensed under a Creative Commons Attribution-Non Commercial-No Derivatives 4.0 International License, cited bySchwartz et al., entitled by: “Genetic Distinctions Between Reticular Pseudodrusen and Drusen: A Genome-Wide Association Study”, American Journal of Ophthalmology, Volume 274, P286-295, June 2025, https://doi.org/10.1016/j.ajo.2025.03.007].
The GWAS analyses reported genetic associations with (i) pure RPD (cases with RPD in at least one eye but no drusen) and (ii) pure drusen (cases with drusen in at least one eye but no RPD). The study included 1,787 participants: 1,037 controls, 361 pure drusen, 66 pure RPD, and 323 mixed cases. The primary pure RPD GWAS identified four genome-wide significant loci: rs11200630 near ARMS2-HTRA1 (P = 1.9e-09), rs79641866 at PARD3B (P = 1.3e-08), rs143184903 near ITPR1 (P = 8.1e-09), and rs76377757 near SLN (P = 4.3e-08). The latter three are uncommon variants (minor allele frequency <5%) and the authors commented that they should be interpreted with caution pending validation in larger studies and external cohorts. A significant association at the CFH locus was also observed using a candidate approach (P = 1.8e-04). In addition, for pure drusen, two loci reached genome-wide significance: rs10801555 at CFH (P = 6.0e-33) and rs61871744 at ARMS2-HTRA1 (P = 4.2e-20).
In summary, the researchers stated that, “we present the first reported GWAS for RPD, finding a genome-wide significant association at the ARMS2-HTRA1 locus, and potentially novel associations at the PARD3B, ITPR1 and SLN loci. An association at CFH was also observed, though at a lower significance threshold, and this locus colocalized with AMD. Larger studies are needed to clarify the roles of ARMS2-HTRA1 and CFH in RPD development and validate these newly identified RPD-specific genetic associations. Future research should aim to quantify RPD burden to enhance the statistical power of these studies.”