Researchers based in the Institute of Ophthalmology, University College London, and Moorfields Eye Hospital, London, UK have presented a deep learning model – “Eye2gene” – capable of providing a genetic diagnosis from a retinal scan. The research study showed a 84% accuracy of a genetic diagnosis from a digital scan, way beyond an evaluation by a human ophthalmic expert. This tool may provide significant supports for patient diagnosis, counselling, patient registries and clinical trial recruitment. In addition, if approved for a wide real-world setting, such tools may potentially become available for remote access with anyone with internet or mobile access. The technical improvement of scans (fundus autofluorescence (FAF), infrared (IR) reflectance imaging and spectral-domain optical coherence tomography (SD-OCT)), and the availability of high-quality training sets in computing, are now likely to diagnose cases far efficiently than previously available.
Rare diseases are a challenge to diagnose and identify the correct pathology, not least of all for the rarity of diagnosticians available in the world. The use of machine learning and AI tools are hugely beneficial to overcome some of these challenges then capable to support clinicians, families and their patients. In a recent study, it was estimated that over 40% of ophthalmic patients in the UK did not obtain a genetic diagnosis, and in many parts of the world where genetic testing is unavailable, showed a further number of undiagnosed cases. To address these concerns, the UCL and Moorfields research teams have developed a deep learning model, (Eye2Gene), now capable to predict the causative IRD gene from the retinal scans of a patient providing a gene-level prediction score for 63 distinct IRD genes. The Eye2Gene model was trained on a total of 58,030 scans from 2,451 patients (4,801 eyes) from Moorfields, split into three different modalities: FAF (n = 16,708), IR (n = 20,659) and SD-OCT volumes (n = 20,663). In addition, the research team asked eight ophthalmologists specializing in inherited retinal degenerations (IRDs), with 5–15 years of experience, to predict the causative gene based on a single FAF image per patient across 50 different patients.
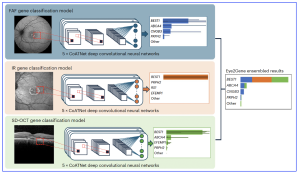
Figure 1. Eye2Gene model. Eye2Gene provides IRD-gene prediction given a retinal scan of one of three imaging modalities (FAF, IR or SD-OCT) for up to 63 gene classes. Images are initially resized to 256 by 256 pixels and rescaled to the range [0,1]. Each image modality-specific predictor block consists of an ensemble of five CoAtNet neural networks. The outputs are averaged to produce a final prediction output. The performance of Eye2Gene is evaluated on a held-out internal test dataset from MEH (Moorfields) consisting of 28,174 images acquired from 524 patients over 9,291 patient visits since 2006. [The research work is licensed under a Creative Commons Attribution-Non Commercial-No Derivatives 4.0 International License, cited by Pontikos N., et al., entitled by: “Next-generation phenotyping of inherited retinal diseases from multimodal imaging with Eye2Gene”, Nature Machine Intelligence, Volume 7, June 2025, 967–978; https://doi.org/10.1038/s42256-025-01040-8].
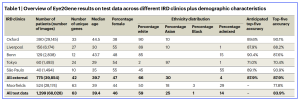
Table 1. Top-five accuracy represents the proportion of case for which the correct gene appeared in the top-five ranked choices of the model. Anticipated top-five accuracy refers to extrapolated accuracy based on per-gene accuracy at MEH extrapolated to target dataset gene distribution. Note that unspecified ethnicity is not accounted in the ethnicity distribution reported here. Bold indicates summary data across sites.
Following their results, their deep learning algorithm, (with an externally validated dataset provided by five different clinical centres) provided better-than-expert-level top-five accuracy of 83.9% for supporting genetic diagnosis for the 63 most common genetic causes. In their study, the UK researchers commented that, ”Eye2Gene shows that next-generation phenotyping using AI is a promising approach to aid in the genetic diagnosis for individuals with IRDs, something that is not only important for improving patient experience and reducing associated overheads, but is likely to become especially important due to the growing number of potentially treatable IRDs where a rapid genetic diagnosis can lead to an improved outcome for the patient.” However, the team also cautioned the outcomes with some caveats including: (1) the training dataset was established in Moorfields Eye Hospital, London, and other gene distributions can vary between different patient populations and across patients of different ethnic backgrounds’; (2) the current Eye2Gene version is limited to predicting 63 gene classes out of a potential of 281 genes associated with IRDs however, there may be other genes that are yet to be confirmed outside of the 63 classes, and finally; (3) the human ophthalmic experts’ evaluation and diagnosis was based on images only, rather than a wider evaluation of clinical decision making and genetic interpretations. Regardless, the learning aspect of the AI model will likely to improve as fine-tuned iterations become updated in due course.