Researchers at the Gavin Herbert Eye Institute, Department of Ophthalmology, University of California, Irvine, has reported an experimental treatment using a CRISPR/Cas9 adenine base editing (ABE) strategy aimed to correct a rhodopsin mutation from an autosomal recessive retinitis pigmentosa (RP) disorder with an in vivo model. Using a subretinal injection with an AAV strategy treatment the study showed rhodopsin correction and restored expression, including partially rescue of retinal function and preservation of retinal structure. Their paper, published in PNAS (2024 Vol. 121 No. 48 e2416827121, https://doi.org/10.1073/pnas.2416827121), showed that “these findings demonstrate that in vivo base editing can restore the function of mutated structural and functional proteins in animal models of disease, including rhodopsin-associated RP and suggest that the timing of gene-editing is a crucial determinant of successful treatment outcomes for degenerative genetic diseases.”
Rhodopsin is a key sensory protein that plays a major structural role in the rod photoreceptor cell and the first genetic mutation linked to retinitis pigmentosa (RP) was the P23H-rhodopsin mutation, causative for autosomal dominant RP (adRP). There are now 150 adRP-causing mutations identified to date, commenting that, “the relative abundance of adRP mutations compared to arRP (autosomal recessive RP) mutations suggests that rhodopsin is highly sensitive to mutation, and mutations in rhodopsin tend to be pathogenic.” One missense mutation studied on the current research, E150K, has shown an autosomal recessive pathology and their researchers have reasoned that “in an autosomal recessive disease such as E150K-arRP, therapeutic rescue could be achieved from reasonably efficient editing, and that in a clinical setting, correction of a subset of rod photoreceptors should arrest the progression of retinal degeneration and provide some benefit to patients.” For CRISPR technology, gene editing has a long history of technical developments aimed to treat genetic disease on the basis of altering a DNA sequence. Antisense oligonucleotides, ribozymes, aptamers, RNA interference (RNAi), zinc-finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs) and CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats). One of the recent versions of gene editing now uses base editing, as a gene therapy approach designed to more finely-tune “surgery”, changing one “letter” of the gene’s code at a time, such as changing a C to T or A to G, aiming to correct the error with a potential promise of more precision, efficiency and safety. Prior to earlier technologies, CRISPR/Cas9 was a significant advance on making alterations on a given specific DNA sequence involving double-strand breaks with a homology-directed repair (HDR) strategy. However, a CRISPR/Cas9 adenine base editing strategy in their current research used dual-adeno-associated viruses encoding an adenine base editor to effect in vivo base editing of rod photoreceptors and demonstrated restoration of wild-type rhodopsin production.
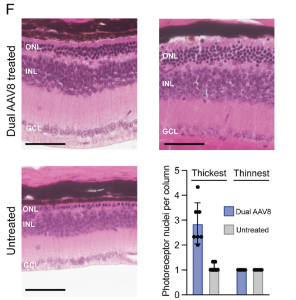
Results of the research showed that rhodopsin could be edited with a maximum of 44% transcript editing from their dual-AAV8 treatment and the outcome was able to rescue the ERG photo-response, compared to untreated controls, which did not show the a- or b-waves in response to photo-stimulation. In addition, the treatment prevented the complete loss of photoreceptor nuclei in the ONL, compared to untreated controls. This led to preservation of the ONL, with a greater number of photoreceptor nuclei per column in the thickest part of each retina (below).
Figure 2. (F) Representative hematoxylin and eosin sections from the thickest region of the retina from dual-AAV8-treated and untreated mice. Quantification of photoreceptor nuclei per ONL, from thickest and thinnest regions of retinas. Scale bar represents 50 μm. ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer. All results are represented as mean ± SD. (This is an open access article, distributed under Creative Commons Attribution License 4.0, authored by Samuel W. Du et al., entitled, In vivo photoreceptor base editing ameliorates rhodopsin-E150K autosomal-recessive retinitis pigmentosa in mice, in PNAS 2024 Vol. 121 No. 48, pp.1-11).
While the researchers commented that similar rare mutations (such as E150K) and other treatment strategies may be difficult to bring a candidate therapy through clinical trials and approval however, other combinations of complementary therapies and / or a simplified regulatory framework could provide RP patients with a molecularly targeted therapy. Professor Jennifer Doudna, a Nobel Prize 2020 winner (Chemistry) for CRISPR, now at the University of California, Berkeley, published a recent paper in Nature (Nature, Oct. 2024, Vol. 634) presenting a “A roadmap for affordable genetic medicines”. While gene therapies require further approvals in the clinic, such as Luxturna, new adaptions will be required across the board, from academe, government, regulation, pharma, venture capital, non-profit initiatives and importantly from patient organisations.